DSI Examples
PENNANT mini-app
PENNANT is an unstructured mesh physics mini-application developed at Los Alamos National Laboratory for advanced architecture research. It contains mesh data structures and a few physics algorithms from radiation hydrodynamics and serves as an example of typical memory access patterns for an HPC simulation code.
This DSI PENNANT example is used to show a common use case: create and query a set of metadata derived from an ensemble of simulation runs. The example GitHub directory includes 10 PENNANT runs using the PENNANT Leblanc test problem.
In the first step, a python script is used to parse the slurm output files and create a CSV (comma separated value) file with the output metadata.
python3 parse_slurm_output.py
In the second step, another python script,
python3 dsi_pennant.py
reads in the CSV file and creates a database:
#!/usr/bin/env python3
"""
This script reads in the csv file created from parse_slurm_output.py.
Then it creates a DSI db from the csv file and performs a query.
"""
import os
from dsi.dsi import DSI
if __name__ == "__main__":
test_name = "leblanc"
table_name = "rundata"
csvpath = f'pennant_{test_name}.csv'
dbpath = f'pennant_{test_name}.db'
datacard = "pennant_oceans11.yml"
output_csv = "pennant_output.csv"
if os.path.exists(dbpath):
os.remove(dbpath)
dsi = DSI(dbpath)
dsi.read(csvpath, "Ensemble", table_name=table_name)
dsi.read(datacard, "Oceans11Datacard")
# saves query output as a Pandas DataFrame to then update that table in the backend
query_output = dsi.query(f"SELECT * FROM {table_name} WHERE hydro_cycle_run_time > 0.006;", collection=True)
dsi.update(query_output)
dsi.write(output_csv, "Csv_Writer", table_name=table_name)
Resulting in the output of the query:
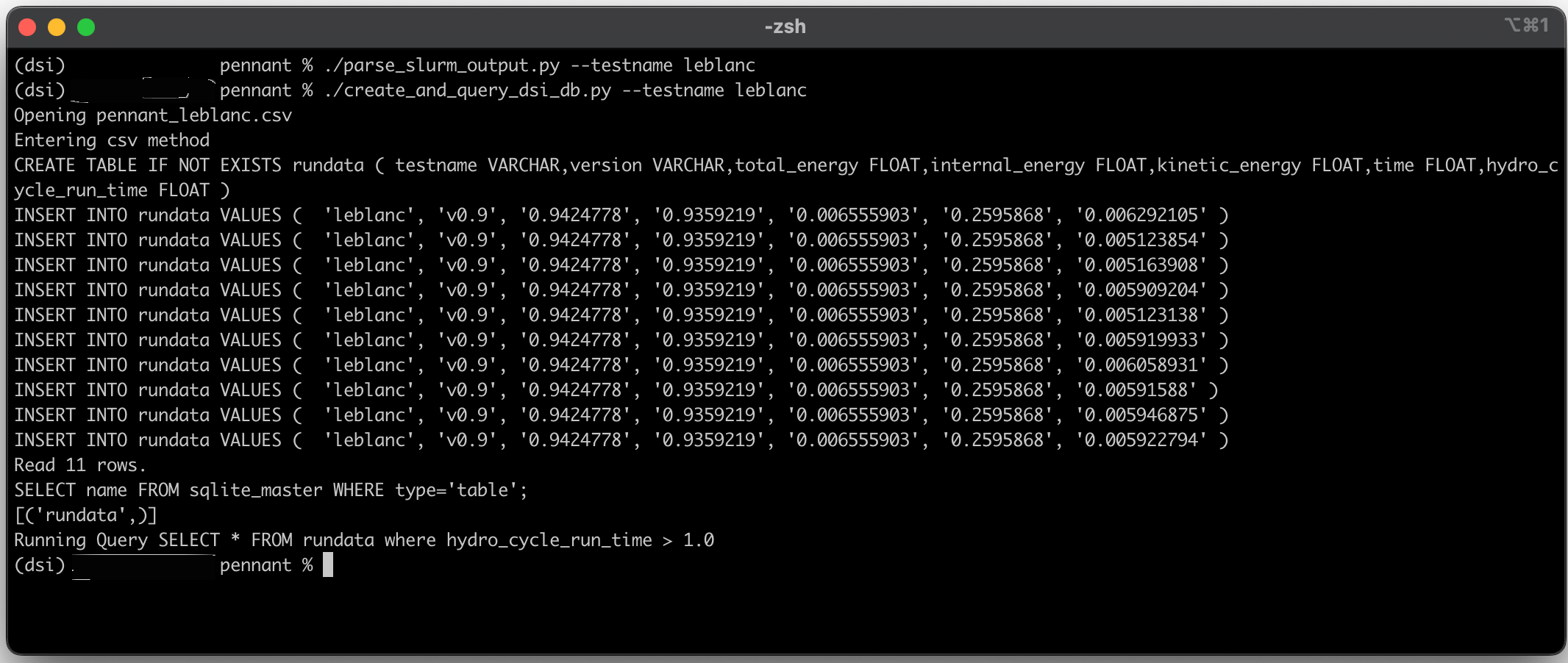
The output of the PENNANT example.
Wildfire Dataset
This example highlights the use of the DSI framework with QUIC-Fire simulation data and resulting images. QUIC-Fire is a fire-atmosphere modeling framework for prescribed fire burn analysis. It is light-weight (able to run on a laptop), allowing scientists to generate ensembles of thousands of simulations in weeks. This QUIC-fire dataset is an ensemble of prescribed fire burns for the Wawona region of Yosemite National Park.
The original file, wildfire.csv, lists 1889 runs of a wildfire simulation. Each row is a unique run with input and output values and associated image url. The columns list the various parameters of interest. The input columns are: wild_speed, wdir (wind direction), smois (surface moisture), fuels, ignition, safe_unsafe_ignition_pattern, safe_unsafe_fire_behavior, does_fire_meet_objectives, and rationale_if_unsafe. The output of the simulation (and post-processing steps) include the burned_area and the url to the wildfire images stored on the San Diego Super Computer.
After loading dsi, run this example within the dsi/examples/wildfire/ folder as all filepaths are relative to that location:
python3 wildfire.py
import os
import pandas as pd
import urllib.request
from dsi.dsi import DSI
def downloadImages(path_to_csv, imageFolder):
"""
Read and download the images from the SDSC server
"""
if not os.path.exists(imageFolder):
os.makedirs(imageFolder)
df = pd.read_csv(path_to_csv)
for url in df["FILE"]:
filename = url.rsplit('/', 1)[1]
dst = imageFolder + filename
if not os.path.exists(dst):
urllib.request.urlretrieve(url, dst)
if __name__ == "__main__":
image_path = "images/"
input_csv = "wildfiredataSmall.csv"
db_name = 'wildfire.db'
output_csv = "wildfire_output.csv"
datacard = "wildfire_oceans11.yml"
table_name = "wfdata"
columns_to_keep = ["wind_speed", "wdir", "smois", "burned_area", "LOCAL_PATH"]
#downloads wildfire images to a local path -- external to DSI
if not os.path.exists(image_path):
os.makedirs(image_path)
downloadImages(input_csv, image_path)
if os.path.exists(db_name):
os.remove(db_name)
dsi = DSI(db_name)
dsi.read(input_csv, "Ensemble", table_name=table_name)
dsi.read(datacard, "Oceans11Datacard")
#get wildfire table's data
wildfire_data = dsi.get_table(table_name, collection=True)
updatedFilePaths = []
for url_image in wildfire_data['FILE']:
image_name = url_image.rsplit('/', 1)[1]
filePath = image_path + image_name
updatedFilePaths.append(filePath)
wildfire_data['LOCAL_PATH'] = updatedFilePaths
#update wildfire table with new column of local paths to the downloaded wildfire images
dsi.update(wildfire_data)
dsi.write(output_csv, "Csv_Writer", table_name=table_name)
dsi.display(table_name, display_cols=columns_to_keep)
Cloverleaf (Complex Schemas)
This example shows how to use DSI with ensemble data from 8 Cloverleaf_Serial runs, and how to create a complex schema compatible with DSI.
The directory with this sample input and output data can be found in examples/clover3d/ where each run has its own subfolder.
Each run’s input file is clover.in and the output is clover.out and the associated VTK files.
After loading dsi, run this example within the dsi/examples/user/ folder as all filepaths are relative to that location:
python3 7.schema.py
This workflow uses a custom Cloverleaf reader to load the data, along with a complex schema that maps the input data, output data, and VTK files to the respective simulation runs. Once executing the workflow, users can see that the state2_density value is the only input parameter changed for each run.
# examples/user/7.schema.py
from dsi.dsi import DSI
schema_dsi = DSI("schema_data.db")
# dsi.schema(filename)
schema_dsi.schema("../test/example_schema.json") # must be before reading Cloverleaf data
#dsi.read(path, reader)
schema_dsi.read("../clover3d/", 'Cloverleaf')
#dsi.write(filename, writer)
schema_dsi.write("clover_er_diagram.png", "ER_Diagram")
#dsi.display(table_name, num_rows, display_cols)
schema_dsi.display("simulation")
schema_dsi.display("input", ["sim_id", "state1_density", "state2_density", "initial_timestep", "end_step"])
schema_dsi.display("output", ["sim_id", "step", "wall_clock", "average_time_per_cell"])
schema_dsi.display("viz_files")
schema_dsi.close()
where examples/test/example_schema.json is:
{
"simulation": {
"primary_key": "sim_id"
},
"input": {
"foreign_key": {
"sim_id": ["simulation", "sim_id"]
}
},
"output": {
"foreign_key": {
"sim_id": ["simulation", "sim_id"]
}
},
"viz_files": {
"foreign_key": {
"sim_id": ["simulation", "sim_id"]
}
}
}
and the generated ER diagram is:
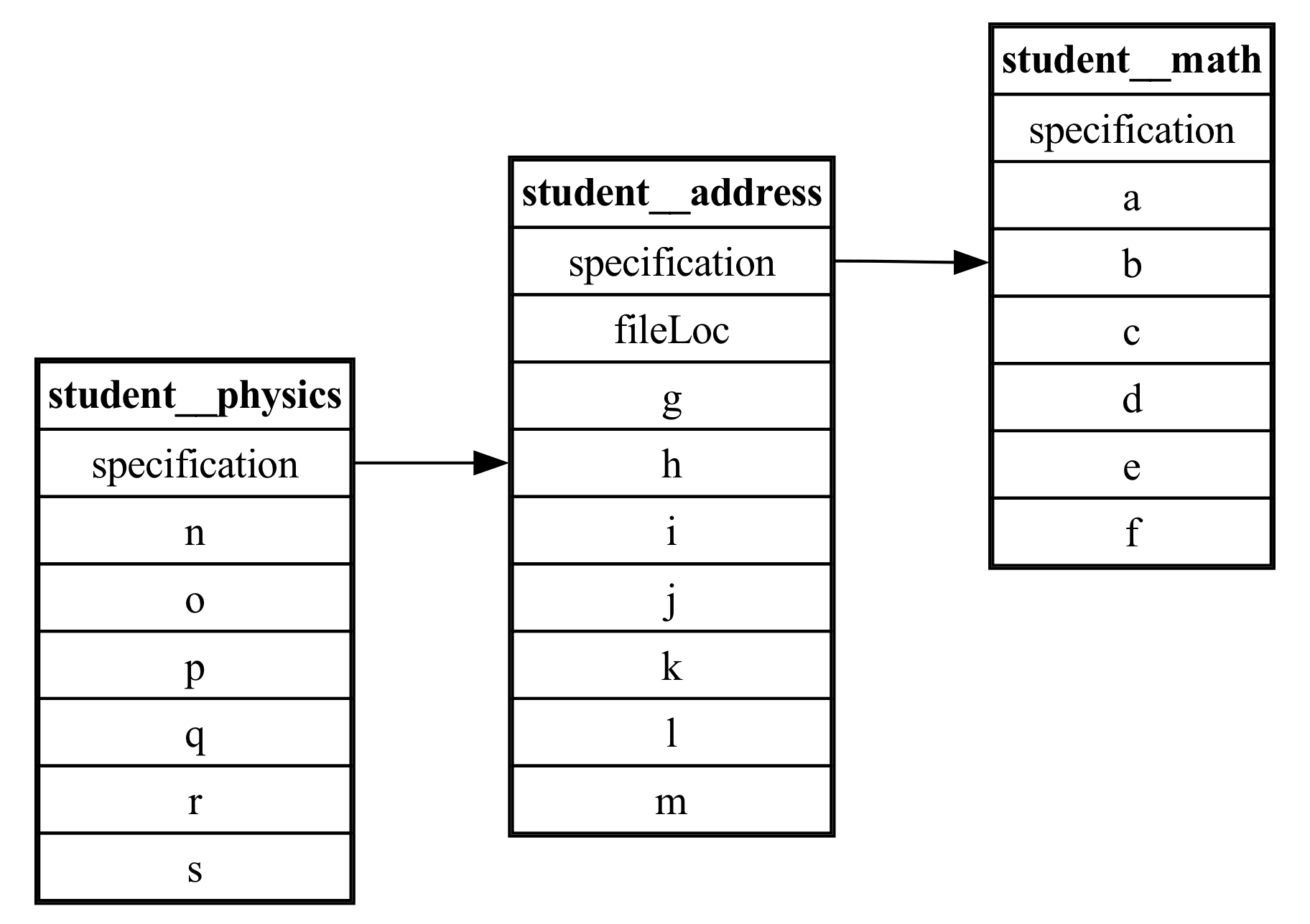
Entity Relationship Diagram of Cloverleaf data. Displays relations between the simulation, input, output, and viz_files tables.
This section explains how to define primary and foreign key relationships in a JSON file for schema(), such as examples/test/example_schema.json
For futher clarity, each schema file must be structured as a dictionary where:
Each table with a relation is a key whose value is a nested dictionary storing primary and foreign key information
Ex from above: “input” : { … }
The nested dictionary has 2 keys: ‘primary_key’ and ‘foreign_key’ which must be spelled exactly the same to be processed:
The value of ‘primary_key’ is this table’s column that is a primary key
Ex from above: “primary_key” : “sim_id”
The value of ‘foreign_key’ is another inner dictionary, since a table can have multiple foreign keys:
Each key in this dictionary is a column in this table that serves as a foreign key
Each value is a list with 2 elements - the table storing the associated primary key, and the column in that table which is the primary key
Ex: “foreign_key” : { “name” : [“table1”, “table1_id”] , “age” : [“table2”, “table2_id”] }
If a table does not have a primary or foreign key, you do not have to include them in the table’s nested dictionary
For example, if we update the Cloverleaf schema by adding a new primary and foreign key relation (assuming the columns exist):
{
"simulation": {
"primary_key": "sim_id"
},
"input": {
"primary_key": "input_id", // <--- new primary key
"foreign_key": {
"sim_id": ["simulation", "sim_id"]
}
},
"output": {
"foreign_key": {
"sim_id": ["simulation", "sim_id"],
"input_id": ["input", "input_id"] // <--- new foreign key
}
},
"viz_files": {
"foreign_key": {
"sim_id": ["simulation", "sim_id"]
}
}
}
our new ER diagram would be:
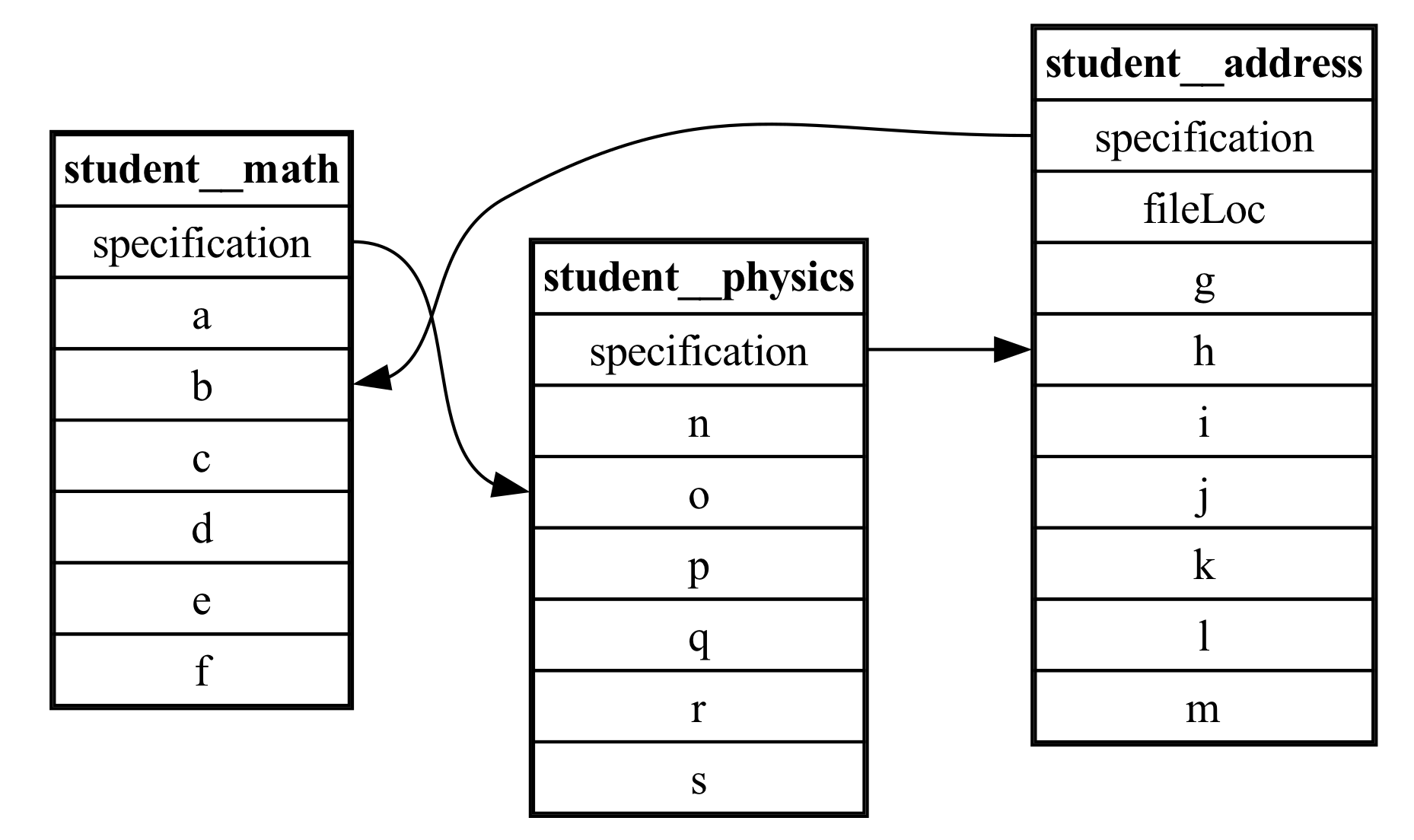
ER Diagram of same data. However, there is now an additional primary/foreign key relation from “input” to “output”