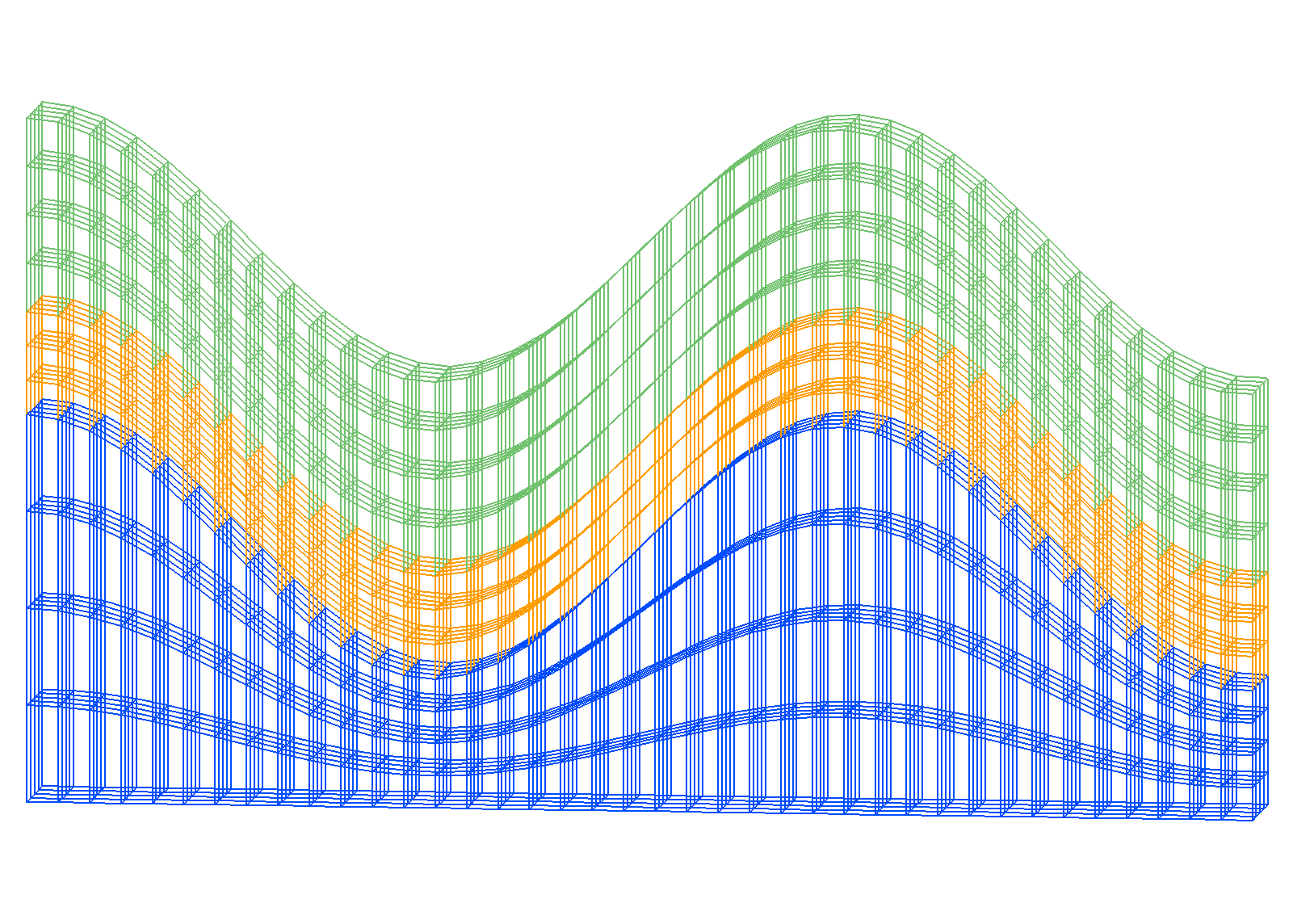
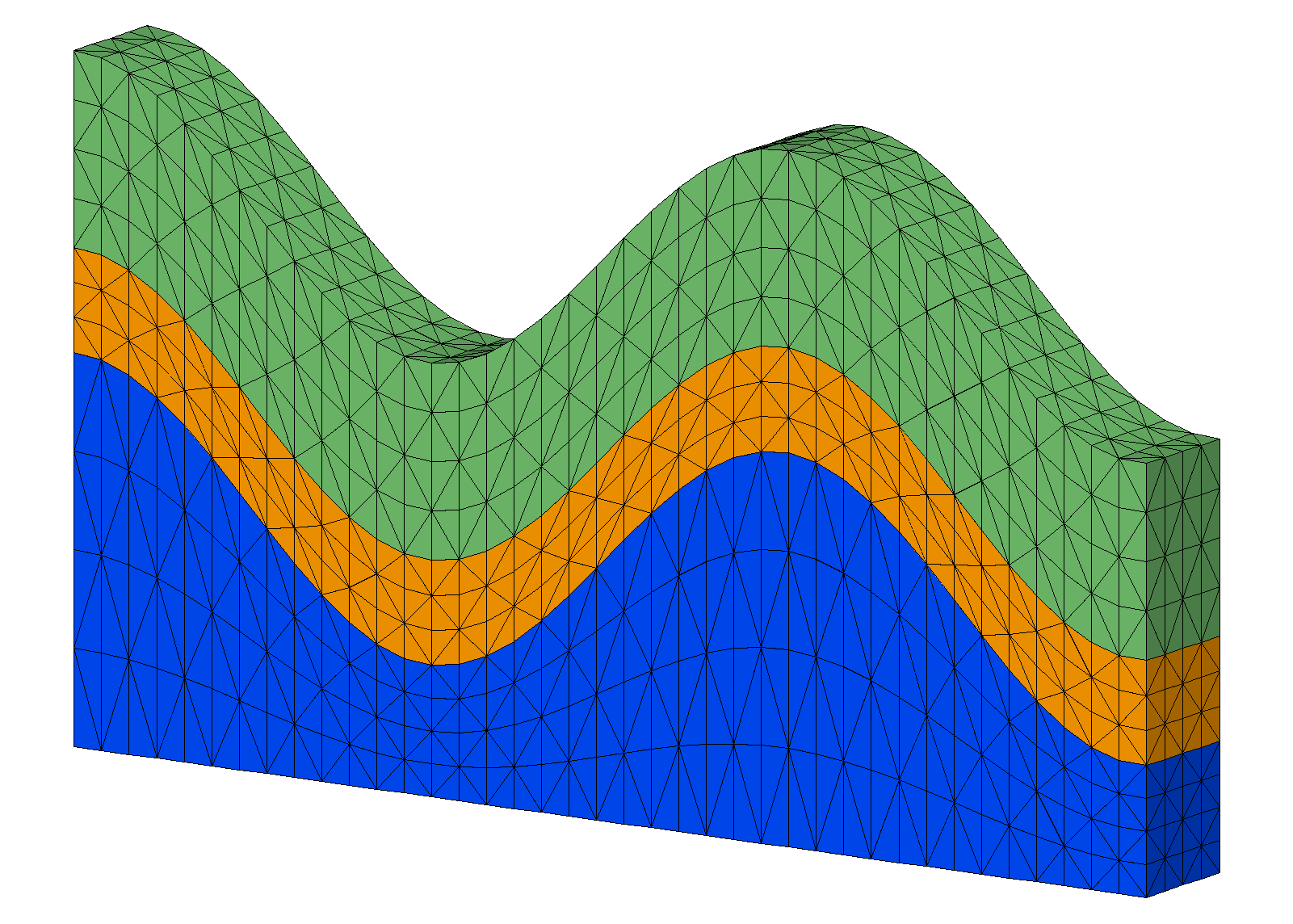
Example: Stacked Mesh using grid2grid
The objective is to stack quad surfaces into a hex mesh then convert each hex element into 5 tets using
grid2grid/hextotet5
Use of hex2tet5 to convert a 3D mesh to a tetrahedral mesh will in general result in a non-Delaunay tetrahedral mesh. If the ultimate goal is a Delaunay mesh, use the connect command, see Examples at connect 3D and connect 2D.
Input Stacked Hex Mesh Output Hex to 5 Tet Mesh
LaGriT command file: grid2grid_wave_tet5.in.txt
LaGriT output report: grid2grid_wave_tet5.out.txt
Steps for this Example:
Step 1. Create hex mesh using createpts and stack/layers
#--* LAGriT Example Input file
#--* Create a non-convex hex and use grid2grid
#----------------------------------------------
# Use x coordinates to compute z based on cos()
# spacing near .25
define / XMAX / 9.5
define / YMAX / 1.
define / NX / 40
define / NY / 5
#-------------------------------------------------
# Create the quad mesh with wave topology
# this is the template used for all surfaces
cmo / create / mosurf / / / quad
quadxy /NX NY/0. 0. 0./ XMAX 0. 0./XMAX YMAX 0./0. YMAX 0./ 1 1 1
createpts/brick/xyz/NX,NY,1/1 0 0 / connect
cmo / setatt / mosurf / imt / 1 0 0 / 1
cmo / setatt / mosurf / itetclr / 1 0 0 / 1
resetpts / itp
math/cos/mosurf/zic/1,0,0/mosurf/xic
dump / surf_wave.inp / mosurf
dump / surf_wave.gmv / mosurf
cmo / printatt / mosurf / -xyz- / minmax
#-------------------------------------------------
# make layers for for input into stack
# top is wave, bottom is flat
define ZBOT 0.
# translate surfaces for vertical spacing
define S1 2.
define S2 .8
define S3 1.5
cmo select mosurf
trans /1,0,0/ 0. 0. 0./ 0. 0. S1
dump / surf_wave_low.inp / mosurf
trans /1,0,0/ 0. 0. 0./ 0. 0. S2
dump / surf_wave_high.inp / mosurf
trans /1,0,0/ 0. 0. 0./ 0. 0. S3
dump / surf_wave_top.inp / mosurf
cmo/setatt/mosurf/ zic/ ZBOT
dump / surf_flat_bot.inp / mosurf
#-------------------------------------------------
# Stack layers from bottom to top wave
cmo/create/mo_stack
stack/layers/avs/ &
surf_flat_bot.inp 1 &
surf_wave_low.inp 2 3 &
surf_wave_high.inp 3 2 &
surf_wave_top.inp 3 3
dump gmv tmp_stack_layers.gmv mo_stack
cmo/printatt/mo_stack/ -xyz- minmax
# connect statcked surfaces into 3D elements
stack/fill/mohex / mo_stack
cmo select mohex
resetpts itp
# write information about this mesh object
cmo/printatt/mohex/-xyz- minmax
quality
# check hex volumes
cmo/addatt/mohex/volume hex_vol
cmo/printatt/mohex/hex_vol minmax
eltset/ ebad / hex_vol/le 0.
dump hex_stack.inp mohex
dump hex_stack.gmv mohex
Step 2. Convert hex elements to tets using grid2grid/hextotet5
#-------------------------------------------------
# convert each hex to 5 tetrahedrals
cmo/select/mohex
grid2grid/ hextotet5/ motet5 / mohex
resetpts/itp
cmo/select/motet5
# report mesh statistics
quality
cmo/addatt/motet5/volume/tet_vol
cmo/addatt/motet5/voronoi_volume/vor_vol
cmo/printatt/motet5/ -all- minmax
finish
Check quality and volume reports for mesh statistics
The mesh statistics show positive volumes for all elements and acceptable aspect ratios. The voronoi volumes for each mesh node are positive but there are many negative coupling coeficients. The impact of these coupling coeficients will depend on the simulation being used.
# report mesh statistics
quality
epsilonl, epsilonaspect: 2.4256785E-12 1.4272489E-35
--------------------------------------------
elements with aspect ratio < .01: 0
elements with aspect ratio b/w .01 and .02: 0
elements with aspect ratio b/w .02 and .05: 0
elements with aspect ratio b/w .05 and .1 : 0
elements with aspect ratio b/w .1 and .2 : 0
elements with aspect ratio b/w .2 and .5 : 2332
elements with aspect ratio b/w .5 and 1. : 6248
min aspect ratio = 0.2881E+00 max aspect ratio = 0.9995E+00
epsilonvol: 1.1179946E-11
---------------------------------------
element volumes b/w 0.2538E-02 and 0.3629E-02: 2672
element volumes b/w 0.3629E-02 and 0.5190E-02: 2992
element volumes b/w 0.5190E-02 and 0.7421E-02: 1572
element volumes b/w 0.7421E-02 and 0.1061E-01: 1056
element volumes b/w 0.1061E-01 and 0.1517E-01: 288
min volume = 2.5381908E-03 max volume = 1.5173843E-02
-----------------------------------------------------------
8580 total elements evaluated.
AMatbld3d_stor: *****Negative Coefficients ******
AMatbld3d_stor: Total Number of Negative Coefficients 4706
AMatbld3d_stor: Number of Significant Negative Coefs 4706
AMatbld3d_stor: Number of 'zero' (< 10e-8 *max) coefs 0
Negative coef at row 2 Column 41 with value -4.1720408E-05
Negative coef at row 2 Column 43 with value -3.5374846E-04
Negative coef at row 2 Column 201 with value -1.8519331E-03
Negative coef at row 2 Column 242 with value -3.8554407E-04
AMatbld3d_stor: npoints = 2400 ncoefs = 26874
AMatbld3d_stor: Number of unique coefs = 14637
AMatbld3d_stor: Maximum num. connections to a node = 19
AMatbld3d_stor: Volume min = 1.6435114E-04
AMatbld3d_stor: Volume max = 5.3887416E-02
AMatbld3d_stor: Total Volume: 4.0775221E+01
AMatbld3d_stor: abs(Aij/xij) min = 0.0000000E+00
AMatbld3d_stor: abs(Aij/xij) max = 8.6219865E-01
AMatbld3d_stor: (Aij/xij) max = 1.9116418E-01
AMatbld3d_stor: (Aij/xij) min = -8.6219865E-01
AMatbld3d_stor Matrix coefficient values stored as scalar area/distance
AMatbld3d_stor Matrix compression used for graph, not coefficient values
motet5 attribute with voronoi volumes created with name vor_vol
cmo/printatt/motet5/-all- minmax
ATTRIBUTE NAME MIN MAX DIFFERENCE LENGTH
-def- 0.000000000E+00 0.000000000E+00 0.000000000E+00 2400
scalar 1 1 0 1
vector 3 3 0 1
nnodes 2400 2400 0 1
nedges 0 0 0 1
nfaces 0 0 0 1
nelements 8580 8580 0 1
mbndry 16000000 16000000 0 1
ndimensions_topo 3 3 0 1
ndimensions_geom 3 3 0 1
nodes_per_element 4 4 0 1
edges_per_element 6 6 0 1
faces_per_element 4 4 0 1
isetwd 0 0 0 2400
ialias 0 0 0 2400
imt1 1 3 2 2400
itp1 0 12 12 2400
icr1 0 0 0 2400
isn1 0 0 0 2400
xic 0.000000000E+00 9.500000000E+00 9.500000000E+00 2400
yic 0.000000000E+00 1.000000000E+00 1.000000000E+00 2400
zic 0.000000000E+00 5.300000000E+00 5.300000000E+00 2400
xtetwd 0 0 0 8580
itetclr 1 3 2 8580
itettyp 5 5 0 8580
itetoff 0 34316 34316 8580
jtetoff 0 34316 34316 8580
itet 1 2400 2399 8580x4
jtet 1 16024947 16024946 8580x4
epsilon 1.000000004E-15 1.000000004E-15 0.000000000E+00 1
epsilonl 2.425678519E-12 2.425678519E-12 0.000000000E+00 1
epsilona 2.649880315E-11 2.649880315E-11 0.000000000E+00 1
epsilonv 1.117994586E-11 1.117994586E-11 0.000000000E+00 1
ipointi 1 1 0 1
ipointj 2400 2400 0 1
idebug 0 0 0 1
itypconv_sm 1 1 0 1
maxiter_sm 25 25 0 1
tolconv_sm 1.000000000E+00 1.000000000E+00 0.000000000E+00 1
nnfreq 1 1 0 1
ivoronoi 1 1 0 1
iopt2to2 2 2 0 1
xmin 0.000000000E+00 0.000000000E+00 0.000000000E+00 1
ymin 0.000000000E+00 0.000000000E+00 0.000000000E+00 1
zmin 0.000000000E+00 0.000000000E+00 0.000000000E+00 1
xmax 9.500000000E+00 9.500000000E+00 0.000000000E+00 1
ymax 1.000000000E+00 1.000000000E+00 0.000000000E+00 1
zmax 5.300000000E+00 5.300000000E+00 0.000000000E+00 1
kdtree_level 0 0 0 1
max_number_sets 64 64 0 1
number_of_psets 0 0 0 1
number_of_eltsets 0 0 0 1
number_of_fsets 0 0 0 1
layertyp -2 2 4 2400
nlayers 12 12 0 1
nnperlayer 200 200 0 1
neperlayer 156 156 0 1
hex_vol 0.000000000E+00 4.552152812E-02 4.552152812E-02 8580
tet_vol 2.538190758E-03 1.517384271E-02 1.263565195E-02 8580
vor_vol 1.643511423E-04 5.388741581E-02 5.372306467E-02 2400
ccoef -1.911641805E-01 0.000000000E+00 1.911641805E-01 2400
ij_ccoef 0 2399 2399 2400