Backends
Backends connect users to DSI Core middleware and allow DSI middleware data structures to read and write to persistent external storage.
Backends are modular to support user contribution, and users are encouraged to offer custom backend abstract classes and backend implementations. A contributed backend abstract class may extend another backend to inherit the properties of the parent.
In order to be compatible with DSI core middleware, backends need to interface with Python built-in data structures and with the Python collections library.
Note that any contributed backends or extensions must include unit tests in backends/tests to demonstrate new Backend capability.
We can not accept pull requests that are not tested.
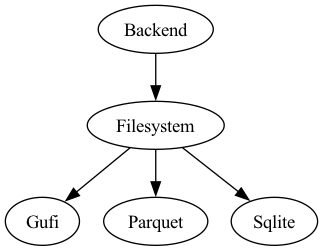
Figure depicts the current DSI backend class hierarchy.
SQLite
- class dsi.backends.sqlite.DataType
Primary DataType Artifact class that stores database schema in memory. A DataType is a generic construct that defines the schema for the tables inside of SQL. Used to execute CREATE TABLE statements.
- class dsi.backends.sqlite.Sqlite(filename, **kwargs)
SQLite Filesystem Backend to which a user can ingest/process data, generate a Jupyter notebook, and find occurences of a search term
- __init__(filename, **kwargs)
Initializes a SQLite backend with a user inputted filename, and creates other internal variables
- close()
Closes the SQLite database’s connection.
- display(table_name, num_rows=25, display_cols=None)
Returns all data from a specified table in this SQLite backend.
- table_namestr
Name of the table to display.
- num_rowsint, optional, default=25
Maximum number of rows to print. If the table contains fewer rows, only those are shown.
- display_colslist of str, optional
List of specific column names to display from the table.
If None (default), all columns are displayed.
- find(query_object)
Searches for all instances of query_object in the SQLite database at the table, column, and cell levels. Includes partial matches as well.
- query_objectint, float, or str
The value to search for across all tables in the backend.
- returnlist
A list of ValueObjects representing matches.
Note: ValueObjects may vary in structure depending on whether the match occurred at the table, column, or cell level.
Refer to find_table(), find_column(), and find_cell() for the specific structure of each ValueObject type.
- find_cell(query_object, row=False)
Finds all cells in the database that match or partially match the given query_object.
- query_objectint, float, or str
The value to search for at the cell level, across all tables in the backend.
- row: bool, optional, default=False
If True, value in the returned ValueObject will be the entire row where a cell matched. If False, value in the returned ValueObject will only be the matching cell value.
return : List of ValueObjects if there is a match.
- ValueObject Structure:
t_name: table name (str)
c_name: list of column names.
If row=True: list of all column names in the table
If row=False: list with one element - the matched column name
value:
If row=True: full row of values
If row=False: value of the matched cell
row_num: row index of the match
type:
If row=True: ‘row’
If row=False: ‘cell’
- find_column(query_object, range=False)
Finds all columns whose names match or partially match the given query_object.
- query_objectstr
The string to search for in column names.
- rangebool, optional, default=False
If True, value in the returned ValueObject will be the [min, max] of the matching numerical column. If False, value in the returned ValueObject will be the full list of column data.
return : List of ValueObjects if there is a match.
- ValueObject Structure:
t_name: table name (str)
c_name: list containing one element - the matching column name
value:
If range=True: [min, max]
If range=False: list of column data
row_num: None
type:
If range=True: ‘range’
If range=False: ‘column’
- find_relation(column_name, relation)
Finds all rows in the first table of the database that satisfy the relation applied to the given column.
- column_namestr
The name of the column to apply the relation to.
- relationstr
The operator and value to apply to the column. Ex: >4, <4, =4, >=4, <=4, ==4, !=4, (4,5), ~4, ~~4
- returnlist of ValueObjects
One ValueObject per matching row in that first table.
- ValueObject Structure:
t_name: table name (str)
c_name: list of all columns in the table
value: full row of values
row_num: row index of the match
type: ‘relation’
- find_table(query_object)
Finds all tables whose names match or partially match the given query_object.
- query_objectstr
The string to search for in table names.
- returnlist of ValueObjects
One ValueObject per matching table.
- ValueObject Structure:
t_name: table name (str)
c_name: list of all columns in the table
value: table data as list of rows (each row is a list)
row_num: None
type: ‘table’
- get_schema()
Returns the structural schema of this database in the form of CREATE TABLE statements.
- return: str
Each table’s CREATE TABLE statement is concatenated into one large string.
- get_table(table_name, dict_return=False)
Retrieves all data from a specified table without requiring knowledge of SQL.
This method is a simplified alternative to query_artifacts() for users who are only familiar with Python.
- table_namestr
Name of the table in the SQLite backend.
- dict_returnbool, optional, default=False
If True, returns the result as an OrderedDict. If False, returns the result as a pandas DataFrame.
- returnpandas.DataFrame or OrderedDict
If dict_return is False: returns a DataFrame
If dict_return is True: returns an OrderedDict
- get_table_names(query)
Extracts all table names from a SQL query. Helper function for query_artifacts() that users do not need to call
- querystr
A SQL query string, typically passed into query_artifacts().
- return: list of str
List of table names referenced in the query.
- ingest_artifacts(collection, isVerbose=False)
Primary function to ingest a collection of tables into the defined SQLite database.
Creates the auto generated runTable if the corresponding flag was set to True when initializing a Core.Terminal Also creates a dsi_units table if any units are associated with the ingested data values.
Can only be called if a SQLite database is loaded as a BACK-WRITE backend. (See core.py for distinction between BACK-READ and BACK-WRITE.)
- collectionOrderedDict
A nested OrderedDict representing multiple tables and their associated data. Each top-level key is a table name, and its value is an OrderedDict of column names and corresponding data lists.
- isVerbosebool, optional, default=False
If True, prints all SQL insert statements during the ingest process for debugging or inspection purposes.
- ingest_table_helper(types, foreign_query=None, isVerbose=False)
Internal use only. Do not call
Helper function to create SQLite table based on a passed in schema.
- typesDataType
- A DataType-derived object that defines:
the table name as a string,
table properties as a dictionary mapping column names to data,
associated units for each column.
- foreign_querystr, optional, default=None
A valid SQL string specifying foreign key constraints to apply to the table.
- isVerbosebool, optional, default=False
If True, prints the CREATE TABLE statements for debugging or inspection.
- list()
Return a list of all tables and their dimensions from this SQLite backend
- notebook(interactive=False)
Generates a Jupyter notebook displaying all the data in the SQLite database.
If multiple tables exist, each is displayed as a separate DataFrame.
If database has table relations, it is stored as a separate dataframe. If database has a units table, each table’s units are stored in its corresponding dataframe attrs variable
interactive: default is False. When set to True, creates an interactive Jupyter notebook, otherwise creates an HTML file.
- num_tables()
Prints number of tables in this backend
- overwrite_table(table_name, collection)
Overwrites specified table(s) in this SQLite backend using the provided Pandas DataFrame(s).
If a relational schema has been previously loaded into the backend, it will be reapplied to the table. Note: This function permanently deletes the existing table and its data, before inserting the new data.
- table_namestr or list
If str, name of the table to overwrite in the backend.
If list, list of all tables to overwrite in the backend
- collectionpandas.DataFrame or list of Pandas.DataFrames
If one item, a DataFrame containing the updated data will be written to the table.
If a list, all DataFrames with updated data will be written to their own table
- process_artifacts(only_units_relations=False)
Reads data from the SQLite database into a nested OrderedDict. Keys are table names, and values are OrderedDicts containing table data.
If the database contains PK/FK relationships, they are stored in a special dsi_relations table.
- only_units_relationsbool, default=False
USERS SHOULD IGNORE THIS FLAG. Used internally by sqlite.py.
- returnOrderedDict
A nested OrderedDict containing all data from the SQLite database.
- query_artifacts(query, isVerbose=False, dict_return=False)
Executes a SQL query on the SQLite backend and returns the result in the specified format dependent on dict_return
- querystr
Must be a SELECT or PRAGMA SQL query. Aggregate functions like COUNT are allowed. If dict_return is True, the query must target a single table and cannot include joins.
- isVerbosebool, optional, default=False
If True, prints the SQL SELECT statements being executed.
- dict_returnbool, optional, default=False
If True, returns the result as an OrderedDict. If False, returns the result as a pandas DataFrame.
- returnpandas.DataFrame or OrderedDict
If dict_return is False: returns a DataFrame
If dict_return is True: returns an OrderedDict
- sql_type(input_list)
Internal use only. Do not call
Evaluates a list and returns the predicted compatible SQLite Type
- input_listlist
A list of values to analyze for type compatibility.
- return: str
A string representing the inferred SQLite data type for the input list.
- summary(table_name=None)
Returns numerical metadata from tables in the first activated backend.
- table_namestr, optional
If specified, only the numerical metadata for that table will be returned as a Pandas DataFrame.
If None (default), metadata for all available tables is returned as a list of Pandas DataFrames.
- summary_helper(table_name)
Internal use only. Do not call
Generates and returns summary metadata for a specific table in the SQLite backend.
- class dsi.backends.sqlite.ValueObject
Data Structure used when returning search results from
find,find_table,find_column,find_cell, orfind_relationt_name: table name
c_name: column name as a list. The length of the list varies based on the find function. Read the description of each one to understand the differences
row_num: row number. Useful when finding a value in find_cell, find_relation, or find (includes results from find_cell)
type: type of match for this specific ValueObject. {table, column, range, cell, row, relation}
DuckDB
- class dsi.backends.duckdb.DataType
Primary DataType Artifact class that stores database schema in memory. A DataType is a generic construct that defines the schema for the tables inside of SQL. Used to execute CREATE TABLE statements.
- class dsi.backends.duckdb.DuckDB(filename)
DuckDB Filesystem Backend to which a user can ingest/process data, generate a Jupyter notebook, and find occurences of a search term
- __init__(filename)
Initializes a DuckDB backend with a user inputted filename, and creates other internal variables
- check_table_relations(tables, relation_dict)
Internal use only. Do not call.
Checks if a user-loaded schema has circular dependencies.
If no circular dependencies are found, returns a list of tables ordered from least dependent to most dependent, suitable for staged ingestion into the DuckDB backend.
Note: This method is intended for internal use only. DSI users should not call this directly.
- tableslist of str
List of table names to ingest into the DuckDB backend.
- relation_dictOrderedDict
An OrderedDict describing table relationships. Structured as the dsi_relations object with primary and foreign keys.
- return: tuple of (has_cycle, ordered_tables)
has_cycle (bool): True if a circular dependency is detected.
ordered_tables (list or None): Ordered list of tables if no cycle is found; None if a circular dependency exists.
- close()
Closes the DuckDB database’s connection.
return: None
- display(table_name, num_rows=25, display_cols=None)
Returns all data from a specified table in this DuckDB backend.
- table_namestr
Name of the table to display.
- num_rowsint, optional, default=25
Maximum number of rows to print. If the table contains fewer rows, only those are shown.
- display_colslist of str, optional
List of specific column names to display from the table.
If None (default), all columns are displayed.
- find(query_object)
Searches for all instances of query_object in the DuckDB database at the table, column, and cell levels. Includes partial matches as well.
- query_objectint, float, or str
The value to search for across all tables in the backend.
- returnlist
A list of ValueObjects representing matches.
Note: ValueObjects may vary in structure depending on whether the match occurred at the table, column, or cell level.
Refer to find_table(), find_column(), and find_cell() for the specific structure of each ValueObject type.
- find_cell(query_object, row=False)
Finds all cells in the database that match or partially match the given query_object.
- query_objectint, float, or str
The value to search for at the cell level, across all tables in the backend.
- row: bool, optional, default=False
If True, value in the returned ValueObject will be the entire row where a cell matched. If False, value in the returned ValueObject will only be the matching cell value.
return : List of ValueObjects if there is a match.
- ValueObject Structure:
t_name: table name (str)
c_name: list of column names.
If row=True: list of all column names in the table
If row=False: list with one element - the matched column name
value:
If row=True: full row of values
If row=False: value of the matched cell
row_num: row index of the match
type:
If row=True: ‘row’
If row=False: ‘cell’
- find_column(query_object, range=False)
Finds all columns whose names match or partially match the given query_object.
- query_objectstr
The string to search for in column names.
- rangebool, optional, default=False
If True, value in the returned ValueObject will be the [min, max] of the matching numerical column. If False, value in the returned ValueObject will be the full list of column data.
return : List of ValueObjects if there is a match.
- ValueObject Structure:
t_name: table name (str)
c_name: list containing one element - the matching column name
value:
If range=True: [min, max]
If range=False: list of column data
row_num: None
type:
If range=True: ‘range’
If range=False: ‘column’
- find_relation(column_name, relation)
Finds all rows in the first table of the database that satisfy the relation applied to the given column.
- column_namestr
The name of the column to apply the relation to.
- relationstr
The operator and value to apply to the column. Ex: >4, <4, =4, >=4, <=4, ==4, !=4, (4,5), ~4, ~~4
- returnlist of ValueObjects
One ValueObject per matching row in that first table.
- ValueObject Structure:
t_name: table name (str)
c_name: list of all columns in the table
value: full row of values
row_num: row index of the match
type: ‘relation’
- find_table(query_object)
Finds all tables whose names match or partially match the given query_object.
- query_objectstr
The string to search for in table names.
- returnlist of ValueObjects
One ValueObject per matching table.
- ValueObject Structure:
t_name: table name (str)
c_name: list of all columns in the table
value: table data as list of rows (each row is a list)
row_num: None
type: ‘table’
- get_schema()
Returns the structural schema of this database in the form of CREATE TABLE statements.
- return: str
Each table’s CREATE TABLE statement is concatenated into one large string.
- get_table(table_name, dict_return=False)
Retrieves all data from a specified table without requiring knowledge of SQL.
This method is a simplified alternative to query_artifacts() for users who are only familiar with Python.
- table_namestr
Name of the table in the DuckDB backend.
- dict_returnbool, optional, default=False
If True, returns the result as an OrderedDict. If False, returns the result as a pandas DataFrame.
- returnpandas.DataFrame or OrderedDict
If dict_return is False: returns a DataFrame
If dict_return is True: returns an OrderedDict
- get_table_names(query)
Extracts all table names from a SQL query. Helper function for query_artifacts() that users do not need to call
- querystr
A SQL query string, typically passed into query_artifacts().
- return: list of str
List of table names referenced in the query.
- ingest_artifacts(collection, isVerbose=False)
Primary function to ingest a collection of tables into the defined DuckDB database.
Creates the auto generated runTable if the corresponding flag was set to True when initializing a Core.Terminal Also creates a dsi_units table if any units are associated with the ingested data values.
Cannot ingest data if it has a complex schema with circular dependencies, ex: A->B->C->A
Can only be called if a DuckDB database is loaded as a BACK-WRITE backend. (See core.py for distinction between BACK-READ and BACK-WRITE.)
- collectionOrderedDict
A nested OrderedDict representing multiple tables and their associated data. Each top-level key is a table name, and its value is an OrderedDict of column names and corresponding data lists.
- isVerbosebool, optional, default=False
If True, prints all SQL insert statements during the ingest process for debugging or inspection purposes.
- ingest_table_helper(types, foreign_query=None, isVerbose=False)
Internal use only. Do not call
Helper function to create DuckDB table based on a passed in schema.
- typesDataType
- A DataType-derived object that defines:
the table name as a string,
table properties as a dictionary mapping column names to data,
associated units for each column.
- foreign_querystr, optional, default=None
A valid SQL string specifying foreign key constraints to apply to the table.
- isVerbosebool, optional, default=False
If True, prints the CREATE TABLE statements for debugging or inspection.
- list()
Return a list of all tables and their dimensions from this DuckDB backend
- num_tables()
Prints number of tables in this backend
- overwrite_table(table_name, collection)
Overwrites specified table(s) in this DuckDB backend using the provided Pandas DataFrame(s).
If a relational schema has been previously loaded into the backend, it will be reapplied to the table. Cannot accept any schemas with circular dependencies.
Note: This function permanently deletes the existing table and its data, before inserting the new data.
- table_namestr or list
If str, name of the table to overwrite in the backend.
If list, list of all tables to overwrite in the backend
- collectionpandas.DataFrame or list of Pandas.DataFrames
If one item, a DataFrame containing the updated data will be written to the table.
If a list, all DataFrames with updated data will be written to their own table
- process_artifacts(only_units_relations=False)
Reads data from the DuckDB database into a nested OrderedDict. Keys are table names, and values are OrderedDicts containing table data.
If the database contains PK/FK relationships, they are stored in a special dsi_relations table.
- only_units_relationsbool, default=False
USERS SHOULD IGNORE THIS FLAG. Used internally by duckdb.py.
- returnOrderedDict
A nested OrderedDict containing all data from the DuckDB database.
- query_artifacts(query, isVerbose=False, dict_return=False)
Executes a SQL query on the DuckDB backend and returns the result in the specified format dependent on dict_return
- querystr
Must be a SELECT or PRAGMA SQL query. Aggregate functions like COUNT are allowed. If dict_return is True, the query must target a single table and cannot include joins.
- isVerbosebool, optional, default=False
If True, prints the SQL SELECT statements being executed.
- dict_returnbool, optional, default=False
If True, returns the result as an OrderedDict. If False, returns the result as a pandas DataFrame.
- returnpandas.DataFrame or OrderedDict
If dict_return is False: returns a DataFrame
If dict_return is True: returns an OrderedDict
- sql_type(input_list)
Internal use only. Do not call
Evaluates a list and returns the predicted compatible DuckDB Type
- input_listlist
A list of values to analyze for type compatibility.
- return: str
A string representing the inferred DuckDB data type for the input list.
- summary(table_name=None)
Returns numerical metadata from tables in the first activated backend.
- table_namestr, optional
If specified, only the numerical metadata for that table will be returned as a Pandas DataFrame.
If None (default), metadata for all available tables is returned as a list of Pandas DataFrames.
- summary_helper(table_name)
Internal use only. Do not call
Generates and returns summary metadata for a specific table in the DuckDB backend.
- class dsi.backends.duckdb.ValueObject
Data Structure used when returning search results from
find,find_table,find_column,find_cell, orfind_relationt_name: table name
c_name: column name as a list. The length of the list varies based on the find function. Read the description of each one to understand the differences
row_num: row number. Useful when finding a value in find_cell, find_relation, or find (includes results from find_cell)
type: type of match for this specific ValueObject. {table, column, range, cell, row, relation}
GUFI
- class dsi.backends.gufi.Gufi(prefix, index, dbfile, table, column, verbose=False)
GUFI Datastore
- __init__(prefix, index, dbfile, table, column, verbose=False)
prefix: prefix to GUFI commands
index: directory with GUFI indexes
dbfile: sqlite db file from DSI
table: table name from the DSI db we want to join on
column: column name from the DSI db to join on
verbose: print debugging statements or not
- query_artifacts(query)
Retrieves GUFI’s metadata joined with a dsi database query: an sql query into the dsi_entries table