Tensorflow Coupon Models
Branched CNNs were developed in tensorflow to estimate the parameterization of a TePla damage model from a coupon experiment. To read more about the coupon experiment, see our 2023 paper.
Table of Contents:
Model Inputs & Outputs
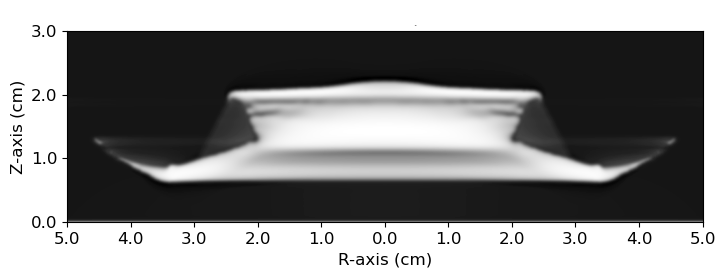
Networks trained on coupon data take as input an image (either a density field or a proton radiograph) and a scalar simulation parameter (a detonation velocity \(D_{cj}\)). An example proton radiograph is shown below.
These models predict values of a parameterized TePla damage model:
\(Y_{spall}\) the spall strength,
\(log_{10}(\phi_0)\) the log base 10 of the initial porosity,
\(\eta\) the overstress parameter,
and the simulation time of an input image \(T_{sim}\).
Running the Coupon Example
To run the scripts on the tensorflow coupon networks, use the following arguments:
--PACKAGE tensorflow
--EXPERIMENT coupon
--MODEL ../examples/tf_coupon/trained_rho2TePla_model.h5or--MODEL ../examples/tf_coupon/trained_pRad2TePla_model.h5
--INPUT_FIELD rhoor--IN_FIELD pRad
--INPUT_NPZuse any .npz file in../examples/tf_coupon/data/
--INPUT_DIR ../examples/tf_coupon/data/
--DESING_FILE ../examples/tf_coupon/coupon_design_file.csv
--NORM_FILE ../examples/tf_coupon/coupon_normalization.npz
--FIXED_KEY tpl112,--FIXED_KEY idx00110, or--FIXED_KEY None
Coupon Data
The file names of coupon data contain multiple pieces of information about their contents. The two relevent componets are the tpl and the idx:
- The tpl is followed by a number identifying a simulation ID.
The values of \(Y_{spall}, log_{10}(\phi_0), \eta\) are identical across files with idential tpl ‘s.
Data is included for tpl112 at all 26 timesteps.
- The idx specifies what time step the simulation was at.
The value of \(T_{sim}\) will be identical across files with identical idx ‘s.
Data is included at idx00110 for 31 differnt simulations.
Each .npz coupon data file contains the following fields:
sim_time (scalar): ground truth for \(T_{sim}\) prediction
pRad (2D array): proton radiograph training field (use
-IN_FIELD pRad)rho (2D array): density training field (use
-IN_FIELD rho)hr_coupon (2D array): density of the coupon
hr_maincharge (2D array): density of the main charge
pressure (2D array)
temperature (2D array)
melt_state (2D array): binary array of if a cell has melted
porosity (2D array)
eqps (2D array): equivalent plastic stress
eqps_rate (2D array): equivalent plastic stress rate
eff_stress (2D array): effective stress
bulk_mod (2D array): bulk modulus of the material
sound_speed (2D array): speed of sound in the material
rVel (2D array): veliocty of material in the R-axis direction
zVel (2D array): velocity of material in the Z-axis direction
Rcoord (1D vector): vector containing position in cm of all cells along the R-axis
Zcoord (1D vector): vector containing position in cm of all cells along the Z-axis
Model Architecture
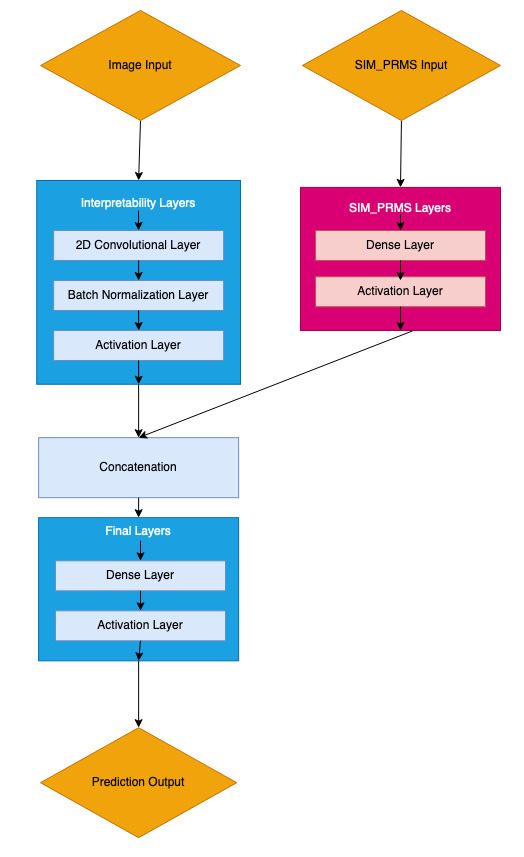
These models have two branches to handle the two disparate input types: an image processing branch, and a simulation parameter processing branch.
Model Layers
The layers in model follow the following naming convention:
???_input: input layer
batch_normalization(_#): batch normalization layer
activation(_#): activation layer
interp_##: 2D convolutional layer that (mostly) preserves size, decreases first two dimensions by 2
second_reduce_##: 2D convolutional layers that more drastically decrease size
dense_?: dense layers
post_reduce_conv: 2D convolutional layer that (mostly) preserves size, decreases first two dimensions by 2
flatten_conv: 2D convolution layer that reduces the first two dimensions to 1
concateate: concatenate layer
final_dense: dense layer that produces outputs
SIM_PRMS_????: Indicates that the layer is on the network branch that operates on the non-image simulation parameter input; layers without this flag are assumed to be on the main network “trunk” that operates on the image input
SIM_PRMS_IMG_???: Indicates the layer where the simulation parameters branch joins the main network “trunk”